Question 1:
Group the following as nitrogenous bases and nucleosides:
Adenine, Cytidine, Thymine, Guanosine, Uracil and cytosine.
Answer:
Nitrogenous base
Adenine
Uracil
Cytosine
Thymine.
Nucleosides
Cytidine
Guanosine
Question 2:
If a double stranded DNA has 20% of cytosines, calculate the per cent of adenine in the DNA.
Answer:
In a double stranded DNA; the amount of A + G = the amount of T + C is equal to one.
Moreover amount of Adenine = amount of thymine
and amount of Guanine = amount of cytosine
Since percentage of cytosine is 20%
∴ percentage of guanine also is 20%
Out of remaining 60%, 30% will be adenine and 30% will be thymine.
Question 3:
If the sequence of one strand of DNA is written as follows: 5'– A T G C A T G C A T G C A T G C A T G C A T G C A T G C –3' Write down the sequence of complementary strand in 5' and 3' direction.
Answer:
5' GCAT GCAT GCAT GCAT GCAT GCAT GCAT 3'
Question 4:
If the sequence of coding strand in a transcription unit is written as follows:
5'– A T G C A T G C A T G C A T G C A T G C A T G C A T G C –3'
Write down the sequence of mRNA.
Answer:
The sequence of mRNA shall be:
5'– U A C G U A C G U A C G U A C G U A C G U A C G U A C G – 3'.
Question 5:
Which property of DNA double helix led Watson and Crick to hypothesize semi-conservative mode of DNA replication, explain.
Answer:
The semi-conservative mode of DNA replication hypothesized by Watson and Crick was based upon the property that during replication the two strands would separate and act as a template for the synthesis of new complementary strands. After the completion of replication, each DNA molecule would have one parental and one newly synthesized strand. This scheme was termed as ‘Semi-conservative’ DNA replication.
Question 6:
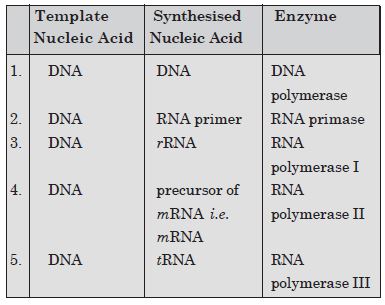
Depending upon the chemical nature of template (DNA or RNA) and the nature of nucleic acids synthesized from it (DNA or RNA), list the types of nucleic acid polymerases.
Answer:
Types of nucleic acid polymerases.
Question 7:
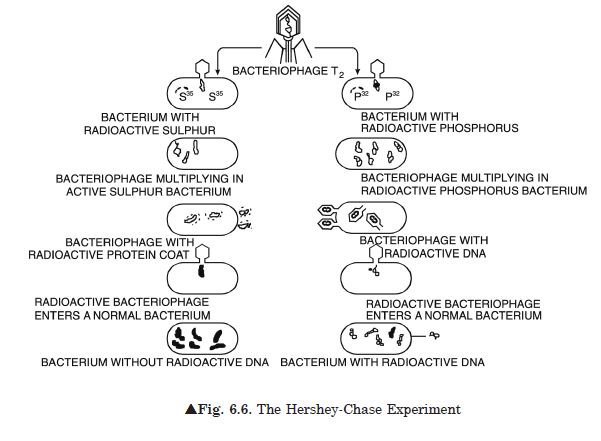
How did Hershey and Chase differentiate between DNA and protein in their experiment while proving that DNA is the genetic material?
Answer:
Alfred Hershey and Martha Chase (1952) worked with viruses that infect bacteria called
bacteriophages.
1. They used radioactive sulphur (35S) to identify protein and radioactive
phosphorus (32P) to identify the components of nucleic acid.
2. The bacteriophage on infection injects only the DNA into the bacterial cell and not the
protein coat; the bacterial cell treats the viral DNA as its own and subsequently
manufactures more virus particles.
3. They made two different preparations of the phage; in one, the DNA was made radioactive
with 32P and in the other, the protein coat was made radioactive with
35S.
4. These two phage preparations were allowed to infect the bacterial cells separately.
5. Soon after infection, the cultures were gently agitated in a blender to separate the
adhering protein coats of the virus from the bacterial cells.
6. The culture was also centrifuged to separate the viral coat and the bacterial cells.
7. Bacteria that were infected with viruses that had radioactive DNA were radioactive,
indicating that DNA was the material that passed from the virus to the bacteria.
8. Bacteria that were infected with viruses that had radioactive proteins were not
radioactive.
9. This indicates that proteins did not enter the bacteria from the viruses.
10. DNA is, therefore, the genetic material that is passed from virus to bacteria.
Question 8:
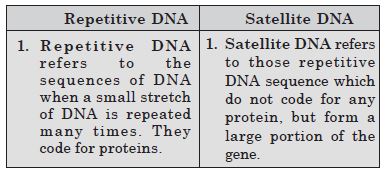
Differentiate between repetitive DNA and satellite DNA.
Answer:
Differences between Repetitive DNA and Satellite DNA
Question 9:
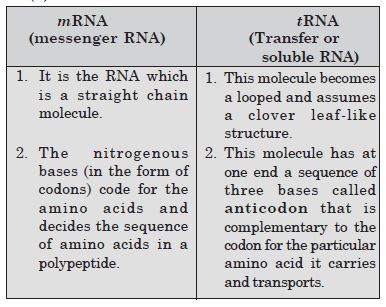
Differentiate between mRNA and tRNA.
Answer:
Differences between mRNA and tRNA
Question 10:
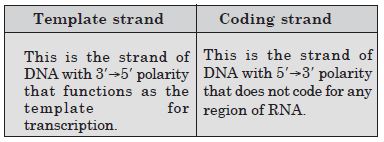
Differentiate between template strand and coding strand.
Answer:
Differences between Template strand and coding strand
Question 11:
Enlist two essential roles for ribosome during translation.
Answer:
Role of ribosomes.
1. Ribosomes usually form linear or helical groups during active protein synthesis called
polyribosomes or polysomes.
2. Ribosome acts as catalyst for formation of peptide bond by an enzyme peptidyl
transferase.
Question 12:
In the medium where E. coli was growing, lactose was added, which induced the lac-operon. But why does lac-operon shut down after some time after addition of lactose in the medium?
Answer:
The addition of lactose induced the lac operon. As lactose formed inducer-repression complex, the operator gene got switched on. As the lactose was digested by the action of enzymes repressor became free and operator gene gets switched off.
Question 13:
Explain (in one or two lines) the function of following:
(a) Promoter
(b) tRNA
(c) Exons
Answer:
(a) Promoter. It is the region of DNA where RNA polymerase attaches prior to the process of
transcription.
(b) tRNA. Works as adaptor molecule for carrying amino acid to the mRNA template during
protein synthesis. It bears anticodon and recognizes the specific
codon on mRNA.
(c) Exons. The region of a gene, which becomes part of mRNA and code for different regions
of the protein are referred to exons.
Question 14:
Why is the Human Genome project called a mega project?
Answer:
1. It is a mega project involving lot of money, most advanced techniques and numerous
computers and scientists at work.
2. Human genome is said to bear 3 × 109 bp. Suppose cost for sequencing one bp is $3 US
dollars, the total cost comes out to be 9 billion US dollars.
3. If such sequences are stored in books, with every page having 1000 letters and each book
is of 1000 pages, total number of books formed will be 3300 to store the information present
in single human cell.
4. Thus, for storing this data, high speed computers are required.
5. Depending upon high magnitude and requirements of the project, HGP has been called as
mega project.
Question 15:
What is DNA fingerprinting? Mention is applications.
Answer:
DNA FINGERPRINTING. Technique of using DNA fragments, resulting from restriction endonuclease
enzyme cleavage to identify particular individuals, is called DNA fingerprinting.
DNA fingerprinting technique was developed by Alec Jeffery (1985, 86) at Leicester
University, United Kingdom. Inheritance of DNA is very stable. Every person has specific
pattern of DNA sequence which shows a
combination of DNA sequence of both mother and father.
Applications of DNA Fingerprinting
1. Paternity disputes can be solved by DNA fingerprinting.
2. It can solve the problems of evolution.
3. It can be used to study the breeding patterns of animals facing the danger of
extinction.
4. It is useful in restoring health of the patients suffering from the leukaemia (blood
cancer).
5. It is very useful in the detection of crime and legal pursuits.
Question 16:
Briefly describe the following:
(a) Transcription
(b) Polymorphism
(c) Translation
(d) Bioinformatics
Answer:
(a) Transcription. Refer to Basis and Basics.
(b) Polymorphism. 1. Polymorphism (variation at genetic level) appears due to mutations. In
simple terms, if inheritable mutation is observed in population at high frequency, it is
referred to as DNA polymorphism.
2. The sequences which do not code for any proteins constitute bulk of human genome. Such
sequences exhibit plenty of polymorphism.
3. DNA from tissues like skin, bone, saliva, sperm, blood, hair follicle represent the same
type of polymorphism is responsible for forming the basis of DNA fingerprinting and Human
Genome Project (HGP).
4. Paternity testing can be carried out by DNA fingerprinting because polymorphism is
transmitted from parents to offspring.
5. Allelic sequence variation is called as DNA polymorphism if one allele or variant at a
locus is found in human population with a frequency higher than 0.01.
6. There is a variety of types of polymorphism, ranging from single nucleotide change to
very large scale change.
7. Such polymorphism plays very important role in speciation and evolution.
(c) Translation. Refer to Basis and Basics.
(d) Bioinformatics. Bioinformatics is a computerassisted interdisciplinary science which
deals with acquisition, storage, management, access and processing of molecular biological
data.
The term Bioinformatics is derived from two words:
‘‘Biology’’ and ‘‘Informatics’’ so bioinformatics concerns the creation and maintenance of
databases of biological informations. It involves the application of computer science and
information technology to analyse and manage biological data. The majority of such databases
are in the form of nucleic acid sequences and the protein sequences derived from them. These
databases are very essential for current and future biotechnology research. In the last few
decades, advances in molecular biology and computer technology have led to rapid sequencing
of large portions of genomes of several species and are responsible for the revolution in
bioinformatics. Thus two main components of bioinformatics are:
1. Computational biology
2. Bioinformatics infrastructure.
Question 17:
Whatever you have learnt from class XI, can you tell whether enzymes are bigger or DNA is bigger in molecular size?
Answer:
The molecular weight of DNA in prokaryotes is 2.6 × 109 (E.coli) and molecular weight of DNA in Eukaryotes is 1.8 × 1012 (humans). While the molecular weight of enzymes varies from 10,000 to one million. Thus, it can be concluded that DNA is bigger in molecular size than enzymes.
Question 18:
What would be the molar concentration of human DNA in a human cell? Consult your teacher.
Answer:
Humans have 3 M of DNA per cell i.e. the molar concentration is 3
